Analysis of SNP and Indel using Resequencing Data
SNPs (Single nucleotide polymorphisms) and small insertion/deletion polymorphisms are the most frequently occurring polymorphisms in the genome, contributing to genetic diversity and phenotypic variation.
When a reference genome is available, resequencing allows for the detection of genomic variations such as SNPs and small insertions/deletions.
When a reference genome is available, resequencing allows for the detection of genomic variations such as SNPs and small insertions/deletions.
Work Flow
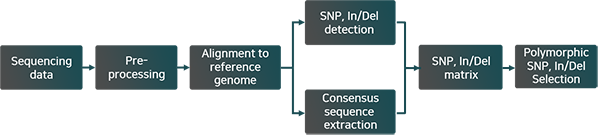
[Genome이 있는 경우 ]
Result Examples
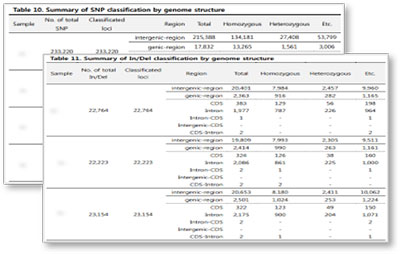 Statistical classification of SNPs(In/Del) based on genome structure
Statistical classification of SNPs(In/Del) based on genome structure
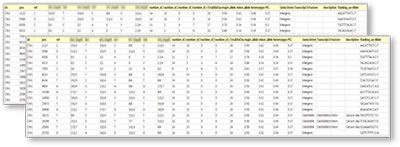 Excel file of SNP analysis results
Excel file of SNP analysis results
Total of 13
related analysis cases
- Exploration of SNP and In/Del markers between samples using RNA data of tomato.
- Exploration of Distinguishing Markers Using Seaweed RNAseq Transcriptome Data
- Exploration of SNP markers between samples related to cold storage using RNA data of sweet potato.
- Exploration of SNP markers between samples using RNA data of shimeji mushroom
- Exploration of SNP and In/Del markers between samples using RNA data of kiwifruit.
- Exploration of SNP and In/Del variations between samples using RNA data of cucumber under cold/stress/disease conditions.
- Exploration of SNP and In/Del markers between samples using RNA data of pepper.
- SNP marker exploration between samples using RNA data of peach.
- Exploration of SNP, In/Del, and SSR markers between samples using RNA data of onions.
- Exploration of SNP and SSR markers between samples using RNA data of cabbage.
- Exploration of SNP, In/Del, and SSR markers between samples using RNA data of ginseng.
- Exploration of SNP variations between samples using RNA data of potatoes.
- SNP marker exploration between samples using RNA data of Miscanthus.
Total of 3


